Abstract
Introduction: Assessment of minimal residual disease (MRD) in a sensitive and timely manner is an essential component of risk stratification in childhood acute lymphoblastic leukemia (ALL). Next generation sequencing (NGS) assays utilize unique genetic sequences created by VDJ rearrangements in leukemia cells to detect MRD at the level of 1 leukemic cell in 1 million cells (Wood et al., 2018). Here we report our experience using NGS MRD for risk group assignment of children and adolescents with newly diagnosed ALL enrolled on the Dana Farber Cancer Institute (DFCI) ALL Consortium Protocol 16-001.
Methods: Patients (pts) ages 1-21 years with B- or T-ALL were eligible for enrollment from 8 centers across the US and Canada. Initial risk status was assigned based on age, presenting leukocyte count, central nervous system (CNS) leukemia status, immunophenotype, and disease biology (Table 1). All patients underwent bone marrow evaluation at diagnosis and again upon completion of remission induction approximately four weeks later (Induction 1a, timepoint 1 (TP1)), with samples evaluated by flow cytometry (FCM) and NGS. NGS was primarily used for MRD-based risk determination, with FCM as a back-up test. Patients with high TP1 MRD (≥10 -4) received intensified therapy and underwent additional MRD assessments at 10 and 20 weeks of therapy.
Multiparametric FCM was conducted locally for 7 of 8 sites in accordance with local CLIA certified lab practices. One site used centralized FCM. NGS MRD was assessed at Adaptive Biotechnologies Corporation, Seattle, WA using the commercially available assay ClonoSEQ ®. Clonality was evaluated at the immunoglobulin (Ig) heavy and light chain (IgH and IgL) and T cell receptor beta and gamma (TCR-B and TCR-G) loci with the maximal sequence used for MRD determination.
Results:
NGS evaluation of MRD is feasible
A total of 317 patients enrolled on 16-001 between 2017 and 2020 were included in this analysis. Among this cohort, NGS identified unique trackable sequences in 98% of pts (N=310). Of the 7 pts without trackable sequences, 57% were pts with early T precursor (ETP) T-ALL (36% of all ETP pts tested). NGS detected trackable sequences in all non-ETP T-ALL pts (N=40), and 99% of B-ALL pts (N=263).
Locus used for MRD determination
Patients with B-ALL had a median of 5 trackable sequences (range 0-14) with 92% having at least one IgH and 64% having at least one TCR-G. For B-ALL, the highest MRD value at TP1 was determined by IGH locus in 44% (N=115) of pts and by TCR-G in 41% (N=109). The IgL or TCR-B locus yielded the highest TP1 MRD value in 15% (N=39). In contrast, pts with T-ALL had fewer trackable sequences with a median of 3 (range 0-8). While 28% (N=13) had at least one Ig sequence, the TCR locus was used for MRD determination in nearly all (98%, N=46) with 94% using TCR-G.
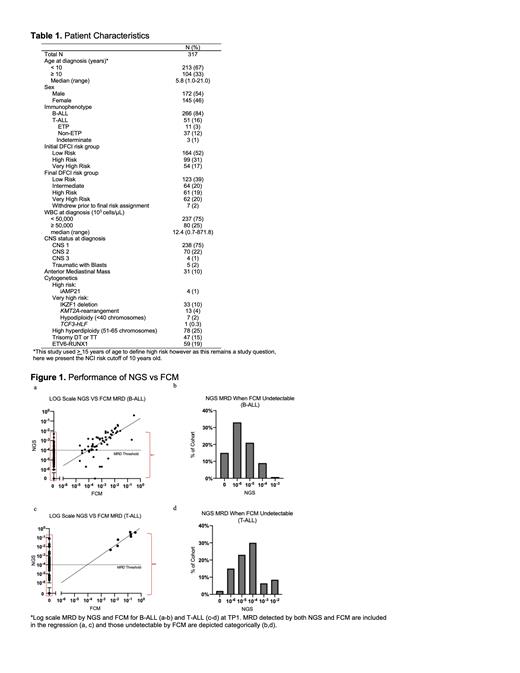
Comparison of NGS and FCM MRD results
NGS and FCM MRD values for 309 pts with results from both assays at TP1 are displayed in Figures 1a-d. Correlation was high between the two modalities for patients with detectable disease by both NGS and FCM (Pearson r=0.87, p<0.0001). NGS additionally detected MRD in the range of 10 -6 to <10 -4 for 160 patients with FCM undetectable disease at TP1, representing 50% of our cohort. Fifty one pts (17%) had high NGS MRD (≥10 -4) but low (8%) or undetectable (92%) FCM MRD (<10 -4), representing 50% of pts classified as high MRD at TP1.
For B-ALL pts with high MRD (N=70), 43% (N=30) were high by NGS (≥10 -4) when FCM was low (<10 -4, N=4) or undetectable (N=26) with 90% of discrepancies at the NGS level of 10 -4 (Figure 1a-b). In contrast, for T-ALL pts with high TP1 MRD (N=28), 75% (N=21) were high by NGS alone, all with undetectable FCM. Sixty seven percent of these pts (N=14) had NGS MRD at the level of 10 -4 and the remaining 33% (N=7) were in the range of 10 -3 to <10 -1 (Figure 1c-d).
Eight pts, all with B-ALL, had low NGS MRD when FCM was above the threshold of 10 -4. One patient had undetectable NGS MRD and the remaining 7 had NGS MRD in the range of 10 -6 to <10 -4.
Conclusions: Delivery of risk adapted therapy for newly diagnosed pediatric pts with ALL utilizing an NGS MRD assay is feasible with evaluable MRD for 98% of patients in our cohort. Importantly, NGS identified more cases as having high MRD than FCM, with the majority of discrepant cases just above the FCM limit of detection (10 -4). NGS provided improved resolution in the range of 10 -6 to <10 -4 for both B-ALL and T-ALL. The prognostic relevance of these low MRD levels awaits longer follow-up.
Kirsch: Adaptive Biotechnologies: Current Employment, Current holder of stock options in a privately-held company. Silverman: Takeda, Servier, Syndax, Jazz Pharmaceuticals: Current equity holder in publicly-traded company, Membership on an entity's Board of Directors or advisory committees.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal